Loading...
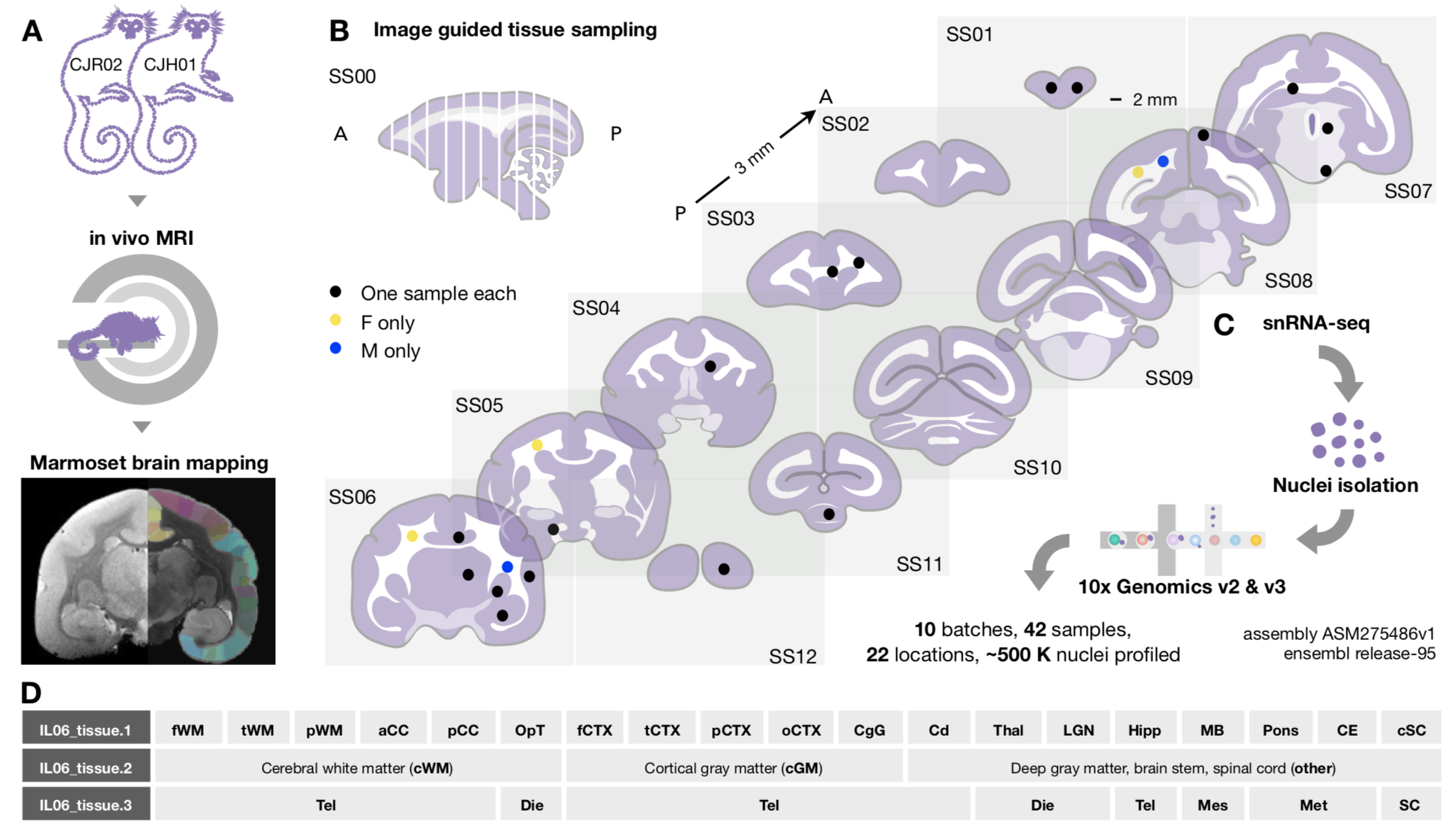 fWM
frontal white matter;
tWM
temporal white matter;
pWM
parietal white matter;
aCC
anterior corpus callosum;
pCC
posterior corpus callosum;
OpT
optic tract;
fCTX
frontal cortex;
tCTX
temporal cortex;
pCTX
parietal cortex;
oCTX
occipital cortex;
CgG
cingulate gyrus;
Cd
caudate;
Thal
thalamus;
LGN
lateral geniculate nucleus;
Hipp
hippo-campus;
MB
midbrain;
Pons
Pons;
CE
cerebellum;
cSC
cervical spinal cord;
Tel
Telencephalon;
Die
Diencephalon;
Mes
Mesencephalon;
Met
Metencephalon.
fWM
frontal white matter;
tWM
temporal white matter;
pWM
parietal white matter;
aCC
anterior corpus callosum;
pCC
posterior corpus callosum;
OpT
optic tract;
fCTX
frontal cortex;
tCTX
temporal cortex;
pCTX
parietal cortex;
oCTX
occipital cortex;
CgG
cingulate gyrus;
Cd
caudate;
Thal
thalamus;
LGN
lateral geniculate nucleus;
Hipp
hippo-campus;
MB
midbrain;
Pons
Pons;
CE
cerebellum;
cSC
cervical spinal cord;
Tel
Telencephalon;
Die
Diencephalon;
Mes
Mesencephalon;
Met
Metencephalon.
Loading...
Callithrix jacchus primate cell atlas (CjPCA):
-
Switch dataset:
Select 1K, MIC, OPC, OLI, AST, VAS, NEU dataset from the left sidebar, wait for the UMAP(2D) tab to load before navigating to other tabs
-
Explore UMAP (3D):
Drag to rotate and zoom, and toggle clusters by clicking the legend
-
Feature expression pattern:
Hit Refresh Gene Selection button to prompt top marker selection for each dataset
Use group.by & split.by in the left sidebar to update results in the relevant tabs
Check VlnPlot.points or FeaturePlot.sort boxes in the left sidebar to show or sort nulcei by feature expression level. Note:Y-axis is ln(gene expression) transformed and ◆ indicate median in VlnPlot
-
DotPlot:
Enter genes in the left sidebar, DotPlots will appear in the ✱ DotPlot (local) + group.by & ✱ DotPlot (global) tabs. Note: Only 50 nuclei per L2 subtype are shown in (global)
-
FeaturePlot & VlnPlot:
Enter features in the left sidebar to see them in the ✦ FeaturePlot & ✦ VlnPlot + group.by tabs
-
DotPlot:
-
Gene markers and modules:
Select multiple genes from the table in the GeneSet tab, then click Submit for ✱ DotPlot button to see results
-
Expression table:
Use average.by to update the table in the ExpMx + average.by tab and download csv
